CHALLENGES AND HOPES IN CRISPR CAS TECHNOLOGY IN FUTURE
SALT Journal of Scientific Research in Healthcare
Volume 1, Issue 2, Page 05-12, Published on 19th August 2021
https://doi.org/10.56735/saltjsrh.ms2101020512
Review Article
*DARLA SRINIVASARAO1, UGRESH CHAUHAN1 AND LABISHETTY SAI CHARAN2
1School of Medical and Allied Science, Division of Medical Lab Technology,
Galgotias University, Greater Noida, India
2School of Medical and Allied Science, Division of Optometry,
Galgotias University, Greater Noida, India
*Corresponding Author:
Mr. Darla Srinivasarao, Assistant Professor, School of Medical and Allied Sciences, Department of Medical Lab Technology, Galgotias University, Greater Noida, India.
ORCID ID: 0000-0002-7366-2529
Email: darla.srinivasa@galgotiasuniversity.edu.in
ABSTRACT
Genome editing is a flourishing tool that tends to be a boon for scientists to change an organism’s DNA. With genome editing technology, scientists can add, remove or alter DNA, resulting in the inactivation of target genes, acquisition of novel genetic traits, and correction of pathogenic gene mutations. Different approaches were developed to edit the genome; one among them is CRISP-Cas which was recently developed. CRISPR associated proteins (CRISPR/Cas) are genome editing systems originated from prokaryotes that have allowed researchers to identify, photograph, alter, and annotate specific DNA and RNA sequences in a variety of living cells. Since using CRISPR/Cas is of low cost with high efficiency and good repeatability with a short cycle, it is used globally for genome editing. In the review, we analysed the utilisation of CRISPR/Cas as therapeutics to cure deadly diseases such as AIDS, Hepatitis B and human papillomavirus infection. Since CRISPR/Cas is cost-effective genome editing, the challenges to be faced in future due to usage of CRISPR/Cas and the areas where it can be utilised were discussed briefly in this review.
Key message: Genetic technology, genome editing, editing tool, CRISPR/Cas, cost-effective, therapeutics.
1.0 INTRODUCTION
The term CRISPR, or the bunched routinely interspaced palindromic changes, was first used by Jansen et al. in 2002 to describe a novel group of redundant DNA successions introduced in the genomes of prokaryotes (1,2,3). It was demonstrated that Streptococcus pyogenes and Streptococcus thermophiles Cas9 can be guided by CRISPR RNAs (crRNAs) to cut objective DNA in vitro through shaping a twofold strand break (DSB). Ishino et al. at first found the CRISPR engineering in the 1980s when they saw a “surprising construction” in the 3′ flanking district of the Escherichia coli IAP gene (4). These underlying perceptions have since been affirmed in different life forms, and presently, the particular spacer areas in the CRISPR loci are perceived to be a sort of immune memory framework to ensure against attacking phage or plasmid DNA. Through this framework, the microscopic organisms can remove a short succession from the attacking DNA and document it in the CRISPR locus, where it tends to be gotten to later by the record.
Later work using CRISPR/Cas loci lacking Staphylococcus aureus changed with the ordinarily utilised Streptococcus pyogenes CRISPR locus has shown that various Cas qualities are significant for the underlying ID and extraction of attacking DNA (5). While the part of the Cas proteins is currently better perceived, their job in CRISPR based obstruction was initially a secret. Underlying investigation of various Cas proteins distinguished homology to known endonuclease spaces, proposing a potential job in presenting viral obstruction through the presentation of DSBs. During the actuation of the CRISPR reaction, the exceptional spacer arrangements are translated into short crRNAs. Garneau et al., was one of the main gatherings to show crRNA worked with Cas proteins to prompt DSBs in attacking DNA (6).
2.0 INTRODUCTION
CRISPR (clustered regularly interspaced short palindromic repeat)/CAS (CRISPR associated) administration are versatile resistant components of numerous microorganisms and archaea to shield themselves from attacking nucleic acids (7-10). Since first applied in quite a while in 2013, the CRISPR /Cas framework, in light of an RNA-guided nuclease, has altered how genome altering is performed (11,12). The aggregation of hereditary transformations in cells after some time prompts disease. What’s more, specific quality changes, for example, driver transformations in TP53, EGFR, KRAS, BRAF, HER2, and MET, can make a cell malignant. Treatment techniques are customarily founded on histological subtypes. Be that as it may, notwithstanding the customary histological characterisations, every malignancy type would now be able to be partitioned into different sub-atomic subtypes that have a pivotal job in the treatment decision-making measure. Each sub-atomic subtype is dealt with distinctively, and clinicians can likewise anticipate treatment results and patient endurance. For example, patients with the cellular breakdown in the lungs with EGFR activating transformations are treated with various sorts of tyrosine kinase inhibitors (TKIs, for example, gefitinib, erlotinib, or afatinib, contingent upon the change. However, protection from the TKIs arises either by DNA transformations or/and metabolic changes. Therefore, treatment methodologies are altered depending on the new sub-atomic mark. Be that as it may, ultimately, the tumour cells do not react to any treatment (13). Along these lines, recognisable proof of new remedial focuses to improve patient endurance and clinical results is critical (14,15). As of late, CRISPR /Cas 9 has altogether impacted the field of atomic science and quality treatment. Muscular tumours are the most well-known kind of tumours, yet less advancement has been made for quality treatment based on treatment contrasted and nonsolid tumours, like leukaemia. Be that as it may, the present circumstance is quickly changing with improvements in CRISPR/CAS9. There is a developing measure of promising preclinical information showing CRISPR /Cas9 to be a compelling apparatus to explicitly target malignancy cells and smother tumour development (16,17,18). Notwithstanding, expanding concerns in regard to the wellbeing of CRISPR/CAS framework because of its latent capacity off-target action and development of CRISPR/CAS -safe break freak infections alongside the trouble in its productive conveyance to each and every infection contaminated cell actually is by all accounts an overwhelming undertaking for the full realisation of this promising antiviral methodology (19,20). In this article, the overall idea regarding the plan and viability approval technique for CRISPR/CAS- based antiviral procedure will initially be presented and assessed. At that point, the current status of the CRISPR/CAS-based antiviral way to deal with control major pathogenic human infections, including human immunodeficiency infection (HIV), hepatitis B infection (HBV), herpes infections, human papillomavirus (HPV), leukaemia and other infections will be summed up straightaway. Lastly, this survey will be finished up with viewing in regards to a likely test for the acknowledgement of CRISPR/CAS-based treatment and prospect for CRISPR/CAS-based antiviral methodology later on.
Figure:1.0 (65).
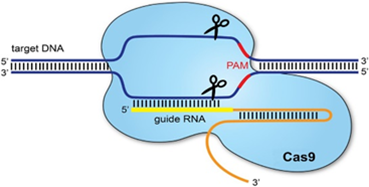
3.0 IMPORTANT THEORY OF PATIENT CURE
3.1 CRISPR/Cas9 therapeutics: a cure for HIV Patient: Current antiretroviral treatment (ART) empowers control of HIV disease at both the individual level and on a worldwide scale. Therefore, consistent decreases have been seen in both quantities of new HIV contaminations just as in HIV death rates (28,29). Two methodologies have been investigated in this specific situation. One depends on the zinc-finger nucleases (ZFNs) that were designed to perceive and separate explicit HIV DNA groupings (30). The second methodology is called TALENS (record activator-like effector nucleases) that program record elements to perceive explicit DNA arrangements (31). The test in the treatment of HIV disease is to beaten the pliancy of the infection. The current helpful arms reservoir comprises of in excess of 25 distinctive antiretroviral intensifies that restrain various strides in the HIV life cycle. At the point when utilised in blend, these antiretroviral medications can significantly control viral replication by keeping prior irregular from reproducing and obtaining extra obstruction changes (32,33,34). Besides, current antiretroviral compounds don’t focus on the incorporated provirus nor smother HIV articulation and creation from the cell repository. Ongoing popular replication or potentially famous creation add to intense irritation, actuation and safe brokenness, causing a wide scope of morbidities in the maturing patient population (35,36). A clinical preliminary with ZFN interceded C-C chemokine receptor type five (CCR5) altering in autologous CD4 T cells had been effectively directed in HIV-1 tainted patients, which illustrated ZFNs-CCR5 adjustment might be powerful and protected in human AIDS treatment (37).
3.2 CRISPR/Cas9 therapeutics: a cure of HBV Virus in Patient: Above 240 million individuals throughout the planet experience the ill effects of persistent HBV disease (38,39). The current HBV treatment system principally depends on nucleoside and nucleotide analogues, which are converse transcriptase inhibitors (40). HBV has a place with the Hepadnaviridae family, including hepatotoxic DNA infections that can taint mammalian and avian hosts. HBV shows the majority of the hereditary designs and replicative attributes seen in other infections in this family (41). A potential test is that HBV DNA can be found in different tissues outside the liver, and the scope of cell lines are lenient for HBV replication (42,43,44). For annihilation of HBV, it is fundamental to convey the nucleases to every single contaminated cell in hepatic and extrahepatic viral supplies. A productive conveyance vehicle ought to be utilised to accomplish the supported enemy of HBV movement in vivo. Because of the low safe potential, non-incorporating nature, and high disease productivity, recombinant adeno-related viral vectors (rAAVs) is right now the ideal decision for conveying CRISPR/Cas9. By and by, the all-out length of the acculturated Cas9 quality (~4.2 kb), sgRNA and the administrative components outperform the ~4.5 kb payload size of rAAV (45).
3.3 CRISPR/Cas9 therapeutics: a cure of HPV virus in the patient: Human papillomaviruses (HPVs) are little DNA infections with a genome size ~8 kb long (Fig. 1). They taint cutaneous or mucosal epithelial cells, genital tissues, and the upper respiratory lot. Until this point, more than 200 hereditarily unmistakable subtypes of HPV have been recognised; also, roughly 90 genotypes have been completely described. Among these sorts, the high-risk HPVs (HR-HPVs), including HPV-16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82, are related with over 90% of cervical malignancies, and less significantly with other anogenital tumours and head and neck malignant growths (46,47,48,49). The HPV genome is made out of an early locale (E) that encodes open perusing outlines associated with the guideline of viral replication and the viral life cycle, what’s more, a late locale (L) that encodes two ORFs (L1 and L2) that structure the viral capsid. Over the span of HR-HPV-interceded malignant growth advancement, the viral DNAs are much of the time coordinated into has cell chromosomes, and the proteins encoded by the viral qualities assume a basic part in carcinogenesis. When contaminating the host, most HPV are cleared in no time; however, some HPVs industriously exist, and viral oncogenes continually express to inactivate p53 and Rb, driving expanded genomic precariousness and aggregation of physical transformations, and now and again incorporation of HPV into the host genome (50,51).
Figure:2.0
4.0 GENOMIC EFFECT OF CRISPR Cas9 THERAPEUTIC
CRISPR (Clustered, regularly interspaced short palindromic repeats) is a guard component of microorganisms and archaea against viral diseases. The presence of these couple changes in the Escherichia coli genome was first described in 1987 and was additionally described two years after the fact (52,53). The CRISPR-intervened versatile insusceptibility in prokaryotes includes three stages: (1) the distinguishing proof of DNA from a pathogenic animal category and resulting joining of the pathogenic arrangement into the prokaryote’s own DNA as a spacer succession, (2) the record furthermore, development of crRNA dependent on that spacer succession, and (3), the crRNA-coordinated cleavage of target pathogenic nucleic acids, affected by means of Cas compound (54,55). Various CRISPR/CAS frameworks (type I, II, III and the more uncommon IV) use particular nucleic corrosive acknowledgement and debasement components (56). CRISPR locus is interpreted to a long RNA particle, known as pre-crRNA, which gets separated, bringing about the creation of crRNA comprised of only one spacer succession. The pre-crRNA cleavage in the most acclaimed CRISPR endogenous hardware, Streptococcus pyogenes, is performed by RNase III and CRISPR-related Csn1 proteins when tracrRNA ties to pre-crRNA due to being totally correlative to the recurrent piece of CRISPR locus coming about in twofold abandoned RNA (57). Recently made crRNA:tracrRNA cross breed next partners with Cas9 protein, creating a functioning ribonucleoprotein (RNP) complex. Cas9 is an RNA-guided endonuclease that can break DNA grouping correlative to the crRNA subunit of the RNP complex. Cas9 restricting site to DNA is a succession named Proto-spacer Adjacent Motif or PAM. The most usually used Cas9 for quality altering is S.pyogenes Cas9 (SpCas9) which perceives a 5′- NGG-3′ PAM site(58,59,60).
Comparison of ZFNs (Zinc Finger Nucleases) and transcription activator-like effector nucleases (TALENs): The previously mentioned instruments have permitted the field of hereditary designing to achieve new statures in site-explicit adjustments; however, both have their own arrangements of upsides and downsides. For example, it is not difficult to plan TALE-DNA restricting exhibits in which every TALE perceives single-base instead of triple base zinc finger proteins. Nonetheless, the cloning of TALE rehashes has been discovered to be more troublesome and, in fact, testing in view of a significant number of indistinguishable recurrent groupings (61). In contrast with ZFNs, TALENs have been discovered to be related to less context-dependent impacts. A setting subordinate impact is seen when the specificities of the solitary fingers rely upon the setting of the adjoining fingers and DNA arrangement. Because of setting reliance, ZFNs show a more significant relationship with the issue of askew impacts. At last, even with various openly accessible strategies for making ZFNs, these nucleases have been developed exclusively by a couple of labs because of troubles related to planning them (62,63).
5.0 CHALLENGES AND OPPORTUNITIES
The significant primary test to the ex vivo quality altering approach is the Conveyance technique. While inspecting the ex vivo CRISPR field in general, electroporation is by and large the most widely recognised strategy to convey CRISPR parts for ex vivo genome altering. In different examinations, viral vectors or manufactured materials (Lipofectamine) were utilised. This addresses one expected freedom for development. Although electroporation can be applied to all cell types and at all phases of cell cycles, its proficiency depends on the cells’ electrical properties, and it has been reported to occasionally cause cell death and content harm (64).
6.0 CONCLUSION
CRISPR/Cas9 genome-altering innovation has prompted the recognisable proof of a few likely remedial focuses in various malignancies. Notwithstanding, the utilisation of CRISPR/Cas9 as a treatment choice in malignant growth includes a few issues, for example, limiting off-target impacts, immunological reactions to Cas9, approval of restorative focuses in creature models, and improvement of the conveyance techniques, which must be tended to prior to going into use in the centre. Regardless of all the previously mentioned difficulties, CRISPR/Cas9 has acquired significant freedoms in the field of malignancy quality treatment and could have a critical part in disease treatment. The CRISPR-Cas framework is an incredible insusceptible framework controlled by prokaryotes against bacteriophages and plasmids. This framework has really altered natural examination in microorganisms as well as in numerous different living beings, possessing its furious streak of advancement, plan straightforwardness, convenience, adaptability, cost adequacy, and effectiveness when contrasted with ZFNs and TALENs. Improved bioinformatics mechanism to recognise the most proper groupings to plan to manage RNAs and streamlining of the trial conditions has empowered extremely powerful methods for abundant presentation of the ideal transformations, initiating extensive energy inside mainstream researchers in saddling the full capability of microbial cells for genome altering, quality guideline, and practical genomics. Up until this point, a few site-explicit changes have been made, which have incredibly helped in understanding the cycles of life and had effectively left an imprint in the field of atomic hereditary qualities. Notwithstanding, there are numerous inquiries that should be tended to. Of these issues, the most significant are the ones that identify with its viability in interlinked quality changes and askew impacts. By the by, ZFNs, TALENs, and particularly the CRISPR-Cas situation are useful assets for controlling the prokaryotic and eukaryotic genomes. Almost certainly, the refinements of these frameworks will proceed, and they will be adjusted in the mix by setting up advancements in better approaches to extraordinarily growing the chances to make new, modern, and continuously important models for essential and translational examination.
REFERENCES
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002 Mar;43(6):1565-75. https://doi.org/10.1046/j.1365-2958.2002.02839.x. PMID: 11952905.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. https://doi.org/10.1126/science.1225829. Epub 2012 Jun 28. PMID: 22745249; PMCID: PMC6286148.
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. https://doi.org/10.1073/pnas.1208507109. Epub 2012 Sep 4. PMID: 22949671; PMCID: PMC3465414.
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987 Dec;169(12):5429-33. https://doi.org/10.1128/jb.169.12.5429-5433.1987. PMID: 3316184; PMCID: PMC213968.
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015 Mar 12;519(7542):199-202. https://doi.org/10.1038/nature14245. Epub 2015 Feb 18. PMID: 25707807; PMCID: PMC4385744.
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010 Nov 4;468(7320):67-71. https://doi.org/10.1038/nature09523. PMID: 21048762.
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709-12. https://doi.org/10.1126/science.1138140. PMID: 17379808.
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010 Jan 8;327(5962):167-70. https://doi.org/10.1126/science.1179555. PMID: 20056882.
- Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012 Dec 20;434(2):202-9.https://doi.org/10.1016/j.virol.2012.10.003. Epub 2012 Nov 2. PMID: 23123013.
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012 Feb 15;482(7385):331-8. https://doi.org/10.1038/nature10886. PMID: 22337052.
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 15;339(6121):819-23. https://doi.org/10.1126/science.1231143. Epub 2013 Jan 3. PMID: 23287718; PMCID: PMC3795411.
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823-6. https://doi.org/10.1126/science.1232033. Epub 2013 Jan 3. PMID: 23287722; PMCID: PMC3712628.
- van der Wekken AJ, Saber A, Hiltermann TJ, Kok K, van den Berg A, Groen HJ. Resistance mechanisms after tyrosine kinase inhibitors afatinib and crizotinib in non-small cell lung cancer, a review of the literature. Crit Rev Oncol Hematol. 2016 Apr;100:107-16. https://doi.org/10.1016/j.critrevonc.2016.01.024. Epub 2016 Jan 25. PMID: 26852079.
- Moore JD. The impact of CRISPR-Cas9 on target identification and validation. Drug Discov Today. 2015 Apr;20(4):450-7. https://doi.org/10.1016/j.drudis.2014.12.016. Epub 2015 Jan 5. PMID: 25572406.
- Ahmad G, Amiji M. Use of CRISPR/Cas9 gene-editing tools for developing models in drug discovery. Drug Discov Today. 2018 Mar;23(3):519-533. https://doi.org/10.1016/j.drudis.2018.01.014. Epub 2018 Jan 8. PMID: 29326075.
- Koo T, Yoon AR, Cho HY, Bae S, Yun CO, Kim JS. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017 Jul 27;45(13):7897-7908. https://doi.org/10.1093/nar/gkx490. PMID: 28575452; PMCID: PMC5570104.
- Floc’h N, Martin MJ, Riess JW, Orme JP, Staniszewska AD, Ménard L, Cuomo ME, O’Neill DJ, Ward RA, Finlay MRV, McKerrecher D, Cheng M, Vang DP, Burich RA, Keck JG, Gandara DR, Mack PC, Cross DAE. Antitumor Activity of Osimertinib, an Irreversible Mutant-Selective EGFR Tyrosine Kinase Inhibitor, in NSCLC Harboring EGFR Exon 20 Insertions. Mol Cancer Ther. 2018 May;17(5):885-896. https://doi.org/10.1158/1535-7163.MCT-17-0758. Epub 2018 Feb 26. PMID: 29483211; PMCID: PMC5932243.
- Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, Yu K, Ruddy DA, Aguirre AJ, Kim JW, Redig AJ, Gainor JF, Williams JA, Asara JM, Doench JG, Janne PA, Shaw AT, McDonald Iii RE, Engelman JA, Stegmeier F, Schlabach MR, Hahn WC. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017 Feb 1; 6: e18970. https://doi.org/10.7554/eLife.18970. Erratum in: Elife. 2017 Oct 31;6: PMID: 28145866; PMCID: PMC5305212.
- Zhu W, Lei R, Le Duff Y, Li J, Guo F, Wainberg MA, Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015; 12:22. https://doi.org/10.1186/s12977-015-0150-z
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013 Mar; 31(3): 230-2. https://doi.org/10.1038/nbt.2507. Epub 2013 Jan 29. PMID: 23360966.
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009 May;10(5):459-66. https://doi.org/10.1016/S1470-2045(09)70025-7. Epub 2009 Mar 9. PMID: 19269895.
- Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016 Jan-Feb;66(1):75-88. https://doi.org/10.3322/caac.21329. Epub 2015 Nov 3. PMID: 26528881; PMCID: PMC4713245.
- Sachdeva M, Sachdeva N, Pal M, Gupta N, Khan IA, Majumdar M, Tiwari A. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015 Nov;22(11):509-17. https://doi.org/10.1038/cgt.2015.54. Epub 2015 Oct 23. PMID: 26494554.
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017 May 6;67(3):177-193. https://doi.org/10.3322/caac.21395. Epub 2017 Mar 1. PMID: 28248415.
- Yang X, Cao W, Wang X, Zhang X, Zhang W, Li Z, Fu H. Down-regulation of 14-3-3zeta reduces proliferation and increases apoptosis in human glioblastoma. Cancer Gene Ther. 2020 Jun;27(6):399-411. https://doi.org/10.1038/s41417-019-0097-7. Epub 2019 May 9. PMID: 31068674.
- Shen Z, Song J, Yung BC, Zhou Z, Wu A, Chen X. Emerging Strategies of Cancer Therapy Based on Ferroptosis. Adv Mater. 2018 Mar;30(12):e1704007. https://doi.org/10.1002/adma.201704007. Epub 2018 Jan 22. PMID: 29356212; PMCID: PMC6377162.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015 Mar;65(2):87-108. https://doi.org/10.3322/caac.21262. Epub 2015 Feb 4. PMID: 25651787.
- Wainberg MA, Zaharatos GJ, Brenner BG. Development of antiretroviral drug resistance. N Engl J Med. 2011 Aug 18;365(7):637-46. https://doi.org/10.1056/NEJMra1004180. PMID: 21848464.
- Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014 Jul 19;384(9939):258-71. https://doi.org/10.1016/S0140-6736(14)60164-1. Epub 2014 Jun 5. Erratum in: Lancet. 2014 Sep 20;384(9948):1098. PMID: 24907868.
- Qu X, Wang P, Ding D, Li L, Wang H, Ma L, Zhou X, Liu S, Lin S, Wang X, Zhang G, Liu S, Liu L, Wang J, Zhang F, Lu D, Zhu H. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013 Sep;41(16):7771-82. https://doi.org/10.1093/nar/gkt571. Epub 2013 Jun 26. PMID: 23804764; PMCID: PMC3763554.
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011 Feb;29(2):143-8. https://doi.org/10.1038/nbt.1755. Epub 2010 Dec 22. PMID: 21179091.
- Götte M. The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance. Curr Opin Virol. 2012 Oct; 2(5):644-50. https://doi.org/10.1016/j.coviro.2012.08.004. Epub 2012 Sep 8. PMID: 22964133.
- Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV Protease Inhibitors: raising the barrier to resistance. Antiviral Res. 2010 Jan; 85(1):59-74. https://doi.org/10.1016/j.antiviral.2009.10.003. Epub 2009 Oct 22. PMID: 19853627.
- Fun A, Wensing AM, Verheyen J, Nijhuis M. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012 Aug 6; 9: 63. https://doi.org/10.1186/1742-4690-9-63. PMID: 22867298; PMCID: PMC3422997.
- Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev. 2013 Jul;254(1):326-42. https://doi.org/10.1111/imr.12065. PMID: 23772629; PMCID: PMC3694608.
- Miedema F, Hazenberg MD, Tesselaar K, van Baarle D, de Boer RJ, Borghans JA. Immune activation and collateral damage in AIDS pathogenesis. Front Immunol. 2013 Sep 26;4:298. https://doi.org/10.3389/fimmu.2013.00298. PMID: 24133492; PMCID: PMC3783946.
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014 Mar 6;370(10):901-10. https://doi.org/10.1056/NEJMoa1300662. PMID: 24597865; PMCID: PMC4084652.
- Lee WM. Hepatitis B virus infection. N Engl J Med. 1997 Dec 11;337(24):1733-45. https://doi.org/10.1056/NEJM199712113372406. PMID: 9392700.
- Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012 Mar 9;30(12):2212-9. https://doi.org/10.1016/j.vaccine.2011.12.116. Epub 2012 Jan 24. PMID: 22273662.
- Block TM, Rawat S, Brosgart CL. Chronic hepatitis B: A wave of new therapies on the horizon. Antiviral Res. 2015 Sep;121:69-81. https://doi.org/10.1016/j.antiviral.2015.06.014. Epub 2015 Jun 22. PMID: 26112647; PMCID: PMC5391532.
- Raimondo G, Caccamo G, Filomia R, Pollicino T. Occult HBV infection. Semin Immunopathol. 2013 Jan;35(1):39-52. https://doi.org/10.1007/s00281-012-0327-7. Epub 2012 Jul 26. PMID: 22829332; PMCID: PMC3540364.
- Dejean A, Lugassy C, Zafrani S, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in pancreas, kidney and skin of two human carriers of the virus. J Gen Virol. 1984 Mar;65 ( Pt 3):651-5. https://doi.org/10.1099/0022-1317-65-3-651. PMID: 6699625.
- Mason A, Wick M, White H, Perrillo R. Hepatitis B virus replication in diverse cell types during chronic hepatitis B virus infection. Hepatology. 1993 Oct;18(4):781-9. https://doi.org/10.1002/hep.1840180406. PMID: 8406351.
- Pontisso P, Poon MC, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in mononuclear blood cells. Br Med J (Clin Res Ed). 1984 May 26;288(6430):1563-6. https://doi.org/10.1136/bmj.288.6430.1563. PMID: 6426645; PMCID: PMC1441279.
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520 (7546): 186-191. https://doi.org/10.1038/nature14299.
- Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F; WHO International Agency for Research on Cancer. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005 Apr;6(4):204. https://doi.org/10.1016/s1470-2045(05)70086-3. PMID: 15830458.
- zur Hausen H. Papillomaviruses in the causation of human cancers – a brief historical account. Virology. 2009 Feb 20;384(2):260-5. https://doi.org/10.1016/j.virol.2008.11.046. Epub 2009 Jan 8. PMID: 19135222.
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004 Jun 20;324(1):17-27. https://doi.org/10.1016/j.virol.2004.03.033. PMID: 15183049.
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017; 543, 378–384. https://doi.org/10.1038/nature21386
- Cain JM, Howett MK. Preventing cervical cancer. Science. 2000 Jun 9; 288(5472): 1753-5. https://doi.org/10.1126/science.288.5472.1753. PMID: 10877694.
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987 Dec; 169(12): 5429-33. https://doi.org/10.1128/jb.169.12.5429-5433.1987. PMID: 3316184; PMCID: PMC213968.
- Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989 Jun;171(6):3553-6. https://doi.org/10.1128/jb.171.6.3553-3556.1989. PMID: 2656660; PMCID: PMC210085.
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014 Nov 28;346(6213):1258096. https://doi.org/10.1126/science.1258096. PMID: 25430774.
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014 Jun 5;157(6):1262-1278. https://doi.org/10.1016/j.cell.2014.05.010. PMID: 24906146; PMCID: PMC4343198.
- Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016 Apr;283(7):1218-31. https://doi.org/10.1111/febs.13586. Epub 2015 Nov 27. PMID: 26535798.
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011 Mar 31;471(7340):602-7. https://doi.org/10.1038/nature09886. PMID: 21455174; PMCID: PMC3070239.
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. https://doi.org/10.1073/pnas.1208507109. Epub 2012 Sep 4. PMID: 22949671; PMCID: PMC3465414.
- Hashemi A, CRISPR–Cas System as a Genome Engineering Platform: Applications in Biomedicine and Biotechnology. Curr. Gene Ther. 2018; 18: 115–124. https://doi.org/10.2174/1566523218666180221110627.
- Sternberg, SH, Redding, S, Jinek, M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507: 62–67. https://doi.org/10.1038/nature13011.
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013; 31(7):397–405. https://doi.org/10.1016/j.tibte ch.2013.04.004.
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014; 32(4):347–355. https://doi.org/10.1038/nbt.2842.
- Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annu Rev Genomics Hum Genet. 2013; 14:135–158. https://doi.org/10.1146/annurev-genom-091212-153435.
- Meacham JM, Durvasula K, Degertekin FL, Fedorov AG. Physical methods for intracellular delivery: practical aspects from laboratory use to industrial-scale processing. J Lab Autom. 2014; 19:1–18.
- Redman M, King A, Watson C, King D. What is CRISPR/Cas9? Arch Dis Child Educ Pract. 2016;101:213-215. http://dx.doi.org/10.1136/archdischild-2016-310459.
ARTICLE TYPE: Review Article;
ORCID ID: Open Researcher and Contributor Identifier (ORCID) ID of corresponding author: https://orcid.org/0000-0002-7366-2529;
ETHICAL: Institutional Ethics Committee;
ACKNOWLEDGEMENT: None;
FINANCIAL DISCLOSURE: The authors declare that there was no financial aid received.;
CONFLICT OF INTEREST: No conflict of interest associated with this review work.;
AUTHORS CONTRIBUTION: All authors discussed the review and prepared the manuscript.;
CORRESPONDING AUTHOR AFFILIATIONS: Mr. Darla Srinivasarao. Assistant Professor, School of Medical and Allied Sciences, Division of Medical Lab Technology, Galgotias University, Greater Noida, India.;
CORRESPONDING AUTHOR EMAIL: darla.srinivasa@galgotiasuniversity.edu; ARTICLE CITATION: Srinivasarao D, Chauhan U, Charan LS. Challenges and hopes in CRISPR CAS technology in future. SALT J Sci Res Healthc. 2021 August 19; 1(2): 05-12.
PUBLISHER’S NOTE: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.



© Darla Srinivasarao, Ugresh Chauhan and Labishetty Sai Charan.
Originally published in the SALT Journal of Scientific Research in Healthcare (https://saltjsrh.in/), 19.08.2021.
This is an open-access article distributed under the terms of the Creative Commons License (https://creativecommons.org/licenses/by-nc-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work, first published in the SALT Journal of Scientific Research in Healthcare (https://saltjsrh.in/), is properly cited. The complete bibliographic information, a link to the original publication on https://saltjsrh.in/, as well as this copyright and license information must be included.
